Emilie Montellier, PhD (1), Nathanaël Lemonnier, PhD (1), Judith Penkert, MD (2), Claire Freycon, MD (1,3,4), Sandrine Blanchet, MSc (1), Amina Amadou, PhD (1,5), Florent Chuffart, PhD (1), Nicholas Fischer, PhD (6), Maria Isabel Achatz, MD, PhD (7), Arnold Levine, PhD (8), Catherine Goudie, MD (4), David Malkin, MD, PhD (6), Gaëlle Bougeard, PhD (9), Christian Kratz, MD (2) and Pierre Hainaut, PhD (1)*
Summary
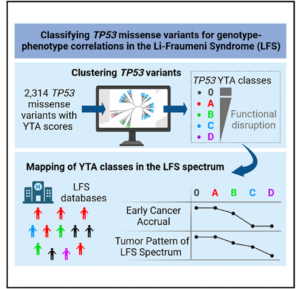
Li-Fraumeni Syndrome is a complex predisposition to multiple cancers over lifetime with presents itself in very different forms from one person and one family to the other. There are many factors that cause these variations but one of them, well recognized, is the fact that TP53 mutations, the genetic defect that causes LFS, are very diverse in their effects on p53 protein function. This has led to the definition of different categories of TP53 variants – pathogenic, likely pathogenic, benign, likely benign – or variants of uncertain significance. In this study, a large international group of researchers and clinicians have pulled together all the data available on the functionalities of thousand of TP53 variants to classify them into 5 robust classes and they show that these classes correspond to different presentations of LFS, from severe to mild/attenuated. This new classification of variants and phenotypes helps to better understand the features of TP53 variants that cause pathogenicity. It will also help clinicians in making cancer risk assessments and decisions, in particular for newly discovered variants that have not yet been associated with LFS.
Affiliations:
(1) Univ. Grenoble Alpes, Inserm 1209, CNRS 5309, Institute for Advanced Biosciences, F38000, Grenoble, France
(2) Pediatric Hematology and Oncology, Hannover Medical School, Hannover, Germany
(3) Centre Hospitalier Universitaire Grenoble Alpes, Department of Pediatrics
(4) Department of Pediatrics, Division of Hematology-Oncology, Montreal Children’s Hospital, McGill University Health Centre, Montreal, Quebec, Canada
(5) Department of Prevention Cancer Environment, Centre Léon Bérard, Lyon, France
(6) Genetics and Genome Biology Program, The Hospital for Sick Children; University of Toronto, Toronto, Ontario, Canada
(7) Department of Oncology, Hospital Sírio-Libanês, São Paulo, Brazil
(8) Simons Center for Systems Biology, Institute for Advanced Study, Princeton, NJ
(9) Department of Genetics, Normandy Center for Genomic and Personalized Medicine, University Hospital, Rouen, France; Normandie Univ – UniRouen, Inserm U1245, Rouen, France
Correspondence:
* corresponding author:
Institute for Advanced Biosciences,
Univ. Grenoble Alpes, Inserm 1209, CNRS 5309
Site Santé, Allée des Alpes
F38700, La Tronche, France
pierre.hainaut@univ-grenoble-alpes.fr
Tel +33 6 20 38 05 47